ExoMethane
Genome-informed Habitability, Biosignature Potential, and Limit of Methane Production in Planetary Environments (ExoMethane)
Faculty Advisor/PI: Dr. Christopher E. Carr (PI), Dr. Rachel A. Moore (Science PI)
Start Date: July, 2022
Current Status: Active
Collaborators: Douglas Bartlett (Co-I, UCSD), Miguel Desmarais (Graduate Student, UCSD), and Jeff Bowman (Collaborator, UCSD)
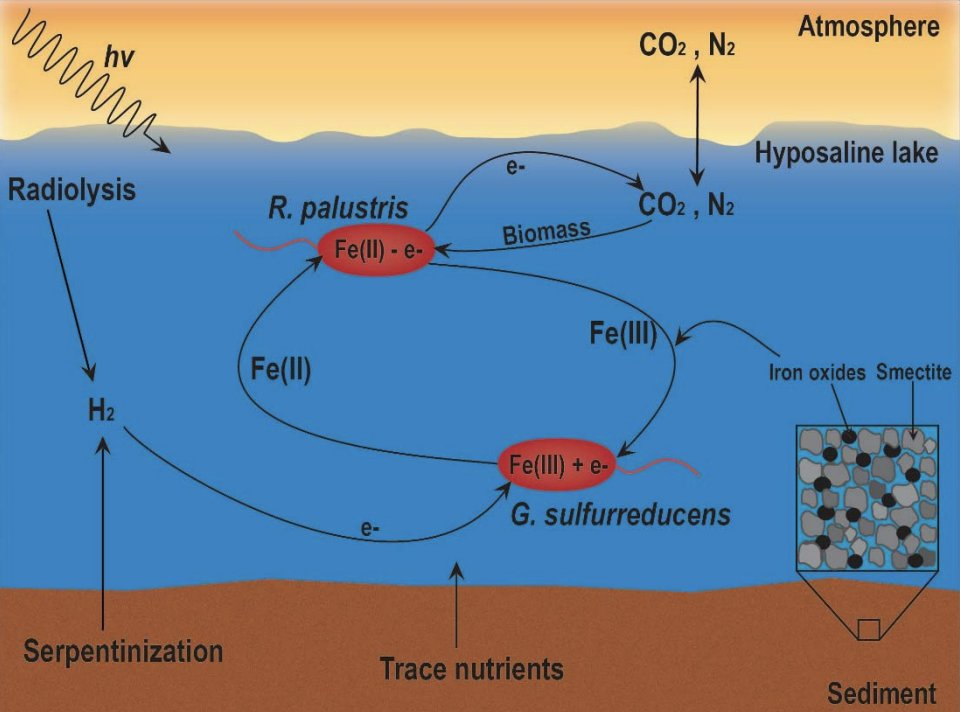
Background: Subsurface environments on Mars and Enceladus are likely presently habitable for life as we know it. Methane measurements by Curiosity and Cassini, respectively, raise the possibility of extant life on these worlds, but at present cannot reliably be distinguished from abiotic sources.
Objectives: Here we propose to leverage both in silico and lab-based experiments to investigate the growth and production of microorganisms in Mars and Enceladus environments. We aim to: 1) Estimate the methanogenic potential of the present subsurface environment of Mars and the ocean on Enceladus, and 2) Determine if microbial methanogenesis can explain the methane observed on Mars and in the plumes of Enceladus.
Methodology: Aim 1a: We will quantify the range of total biomass and biosignatures that could be produced by methanogens given the conditions analyzed by Curiosity and Cassini using genome-scale modeling; we previously used this approach to demonstrate that Gale Lake was once habitable and that detectable levels of biomarkers could have been produced if life was present. Here we expand this work to evaluate habitability based on subsurface conditions and methanogenesis, and predict potential production of a comprehensive set of biosignatures. Aim 1b: We will validate the models with in vitro experimentation of candidate methanogens via long-duration culturing under high-pressure in-situ like conditions. Aim 2a: We will model the range of potential abiotic methane production using a similar framework to aim 1a. Aim 2b: determine if the methane produced in these environments is consistent with abiotic and/or biotic mechanisms by comparing predictions of aim 1a and 2a, and will also identify potential byproducts that could be used in situ to differentiate between abiotic production and extant life.
Relevance: Our work will help to systematically constrain the habitability and biosignature potential of Mars and Enceladus. Of significance, this work would reveal the cell abundance limits sustainable in the subsurface of Mars and Enceladus’ ocean, elucidate limits of biosignature production on Mars and Enceladus, and determine if observed CH4 signals are congruent with a biotic or abiotic source. Our work is responsive to the solicitation themes Biosignatures and Life Elsewhere (primary) and Early Evolution of Life and the Biosphere (secondary). Impacts may include 1) Refining the limits of microbial life; 2) Improving the context of specific biomarkers; 3) Informing future life detection missions and planetary protection.
Funding: NASA Exobiology award # 80NSSC22K0974 to C.E.C. (PI) and R.A.M. (Science PI).
Preprint: Rachel A. Moore, Christopher E. Carr. Microbial habitability of the early Mars lacustrine environment sustained by iron redox cycling. bioRxiv 2021.09.07.459191; doi: https://doi.org/10.1101/2021.09.07.459191